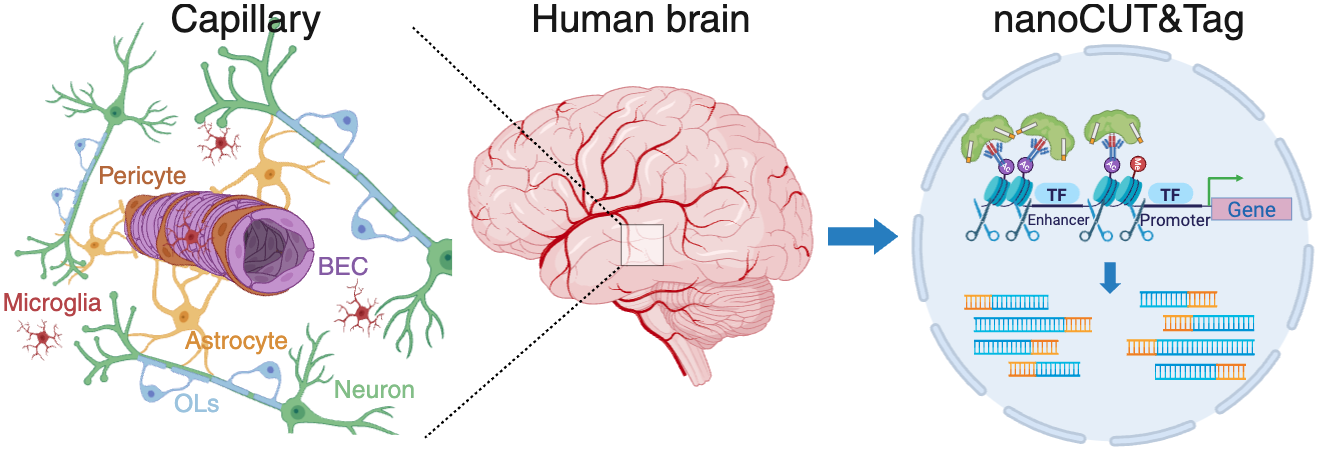
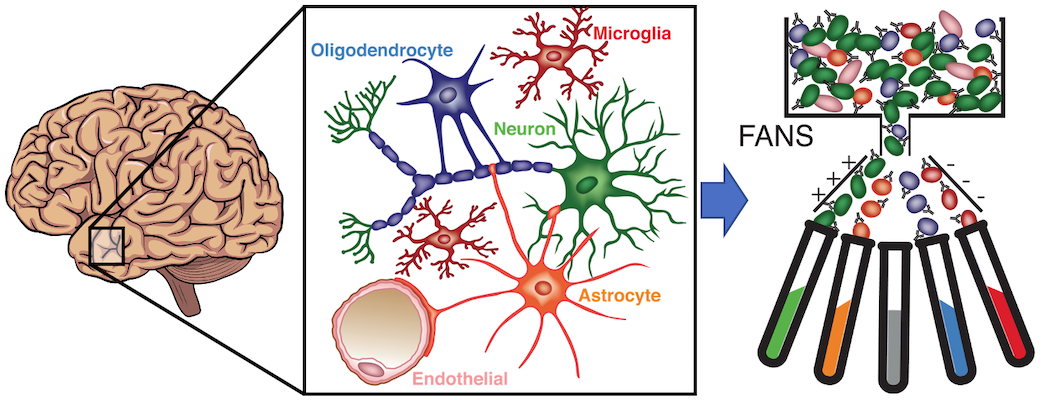
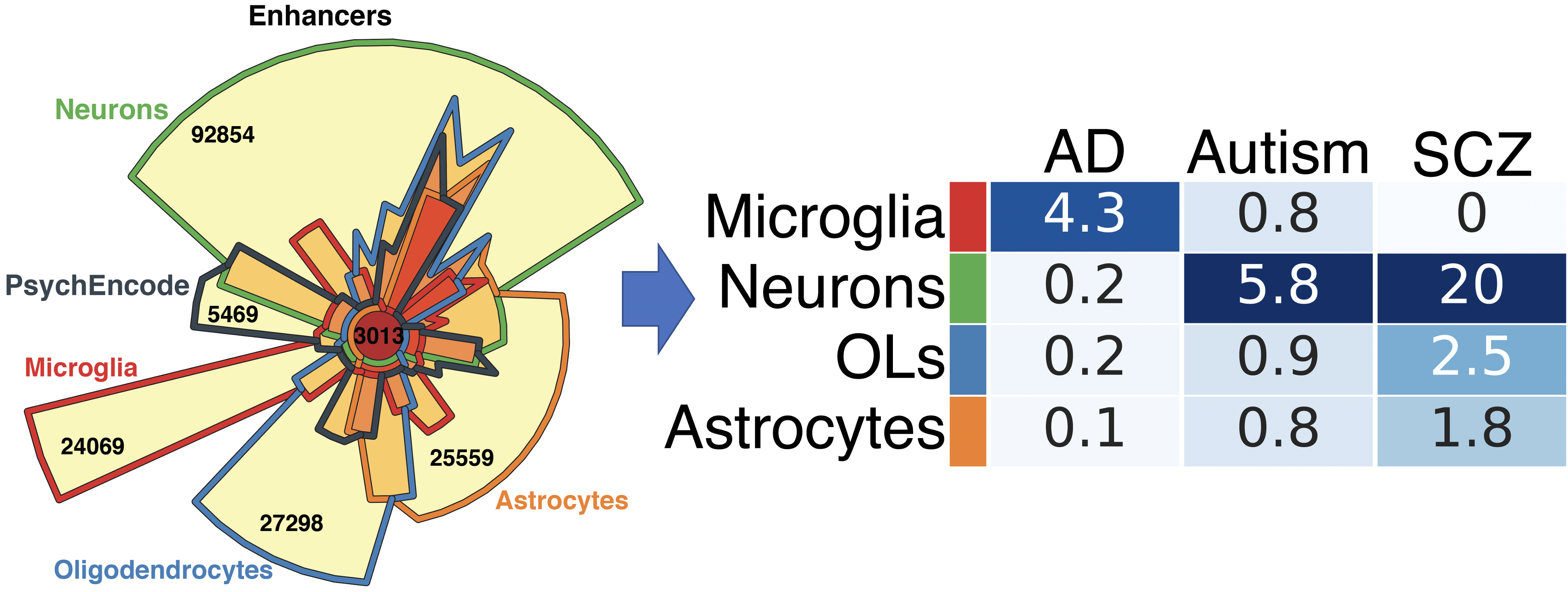
Datasets and resources
- Ziegler*, K.C., Askarova*, A., et al. (2026) Neuron 114(2):268-286.
- Visualization of H3K4me3 and H3K27ac CUTTag datasets and pcHi-C interactomes are available as a UCSC genome browser session
- CUTTag fastq files from postmortem samples and pcHiC fastq files for primary BECs and primary perciytes are available through GEO
- CUTTag and pcHiC fastq files from resected samples are available through European Genome-phenome Archive
- Processed CUTTag peak files and pcHi-C interactome files for each brain cell type are available on GitHub
- Nott*,§, A., Schlachetzki*,§, J.C.M., Fixsen, B.R., Glass§, C.K. (2021) Nat Protoc. 366(6469):1134-1139.
- A full-text view-only version of the protocol is accessible with the following SharedIt link
- Nott*, A., Holtman*, I.R., Coufal*, N.G., et al. (2019) Science 366(6469):1134-1139.
- Visualization of ATAC-seq, ChIP-seq, and PLAC-seq datasets is available as a UCSC genome browser session
- ATAC-seq, ChIP-seq, and PLAC-seq sequencing datasets (fastq files) are available through dbGap
- Processed ATAC-seq and ChIP-seq peak files for each brain cell type are available on GitHub